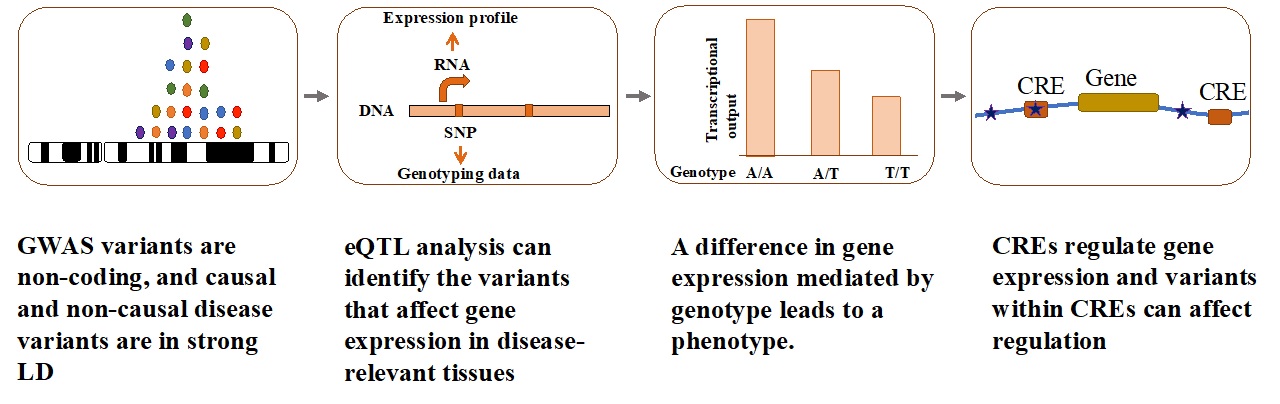
Genome-wide association studies (GWAS) have established a major role of non-coding variants in Age-related macular degeneration (AMD), which is the leading cause of blindness in the elderly. However, the biological interpretation of AMD-GWAS findings remains challenging as we have a limited understanding of how associated non-coding variants translate into disease pathogenesis. In recent years, gene expression regulation has emerged as a dominant mechanism for non-coding variants to mediate the disease risk. As a postdoc , I aided functional interpretation of AMD-associated, non-coding variants by performing a large scale, integrative analyses of transcriptome (RNAseq) and their genetic variations that led to generation of a reference for expression quantitative trait loci (eQTL) in human retina that was missing from Genotype-Tissue Expression (GTEx) and other large datasets. Here at Baylor College of Medicine, we are extending this work by generating and integrating multiple high-throughput functional genomics data sets on the genome, transcriptome, and epigenome, in disease-relevant tissues/cells to identify causal variant, disease-associated cis-regulatory regions (CREs), target genes, and the disease mechanism underlying GWAS loci.
Related publications
- Ratnapriya R, et al.,. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019;51:606-10.
- Fritsche et al., A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134-43.
- Ratnapriya et al., Genetic studies of Age-related macular degeneration: lessons, challenges and opportunities for disease management. Ophthalmology. 2012;119:2526-36.
- Strunz T, et al., .A mega-analysis of expression quantitative trait loci in retinal tissue. PLoS Genet. 2020;16:e1008934.