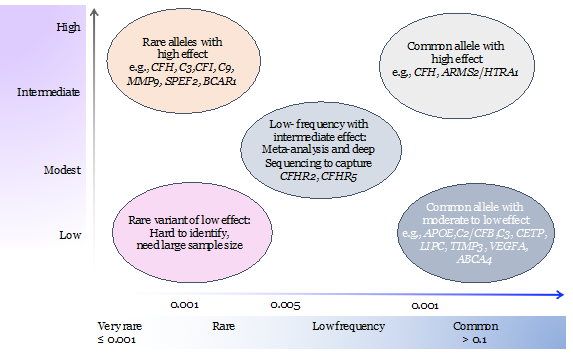
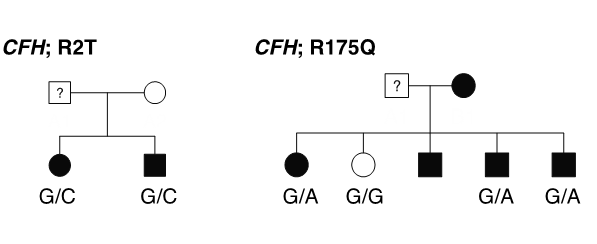
GWAS signals are primarily, common, non-coding variants and identity of the causal gene is not obvious . In a shift from common-disease common-variants (CDCV) hypothesis, the role of rare variants are being recognized in complex diseases. Identification of a rare variants can provide hypothesis-free evidence for causality by pointing to the target gene underlying AMD. Thus, we apply whole exome sequencing to identify the rare variants in AMD and related maculopathies. Our most recent work on the analysis of 63 multiplex AMD families identified rare variants and target genes at eight AMD loci.
Related publications
- Ratnapriya et al., Family-based exome sequencing identifies rare coding variants in age-related macular degeneration. Hum Mol Genet. 2020;29:2022-34.
- Ratnapriya et al., Rare and common variants in extracellular matrix gene Fibrillin 2 (FBN2) are associated with macular degeneration. Hum Mol Genet 2014;23:5827-37.
- Zhan et al., Targeted sequencing, augmented with public resources, identifies a rare Complement 3 allele associated with AMD. Nat Genet. 2013;45:1375-9.